Installation and hello world of rHadoop
Recently I needed to install and configure a local version of rHadoop.
I ended up using the Revolution Analytics rHadoop packages, running inside RStudio server on a CentOS based Cloudera quickstart VM.
Here are instructions based on a couple of sites: ashokharnal’s tutorial and Imre Kocsis’s tutorial.
The exact instructions didn’t quite work for me (java problems), so here is what I did.
Set up VirtualBox and the Cloudera image
First download and install VirtualBox to run your VM.
Next, get a copy of the Cloudera quickstart VM. It’s about 4GB and needs unzipping and copying to run, so plan accordingly.
Once you have virtualbox installed, import the image, using File>Import Appliance.
Default settings are ok, but you will want to change network setting so you can access RStudio server from your native OS. My network is easy - on the settings panel, simply choose network and set attached to: bridged.
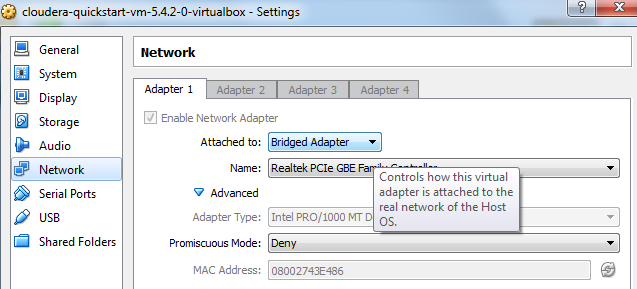
Note: Be careful of network security - if you aren’t careful your VM will be accessible to the world.
Now boot the VM and wait for it to start up. It will take a while at the boot screen.

Get and install R and dependencies
Once cloudera boots, open the console, and install R and R-devel (my version of cloudera came with EPEL repos enabled, see here for instruction if not)
$ sudo yum install R R-develNow we need to install various packages in R that the hadoop packages depend on
Boot R:
$ RThen inside R, install the packages and then quit
> install.packages(c("RJSONIO","Rcpp", "bitops", "caTools", "digest",
"functional", "stringr", "plyr", "reshape2","rJava"))
> q()Get and install rHadoop packages
Now we want to download the rHadoop packages. For now, I only want to do some map reducing, so only need rmr2 and rhdfs.
The packages aren’t on CRAN, but are available on GitHub
I used version 3.3.1 of rmr2 and version 1.0.8 of rhdfs
Note: You don’t want the windows version if you are using cloudera.
Before installing, we must set up some environment variables to allow R to see Hadoop.
Add the lines:
export HADOOP_CMD=/usr/bin/hadoop
export HADOOP_STREAMING=/usr/lib/hadoop-0.20-mapreduce/contrib/streaming/hadoop-streaming-2.6.0-mr1-cdh5.4.2.jarto both ~/.bashrc and /etc/profile (eg sudo nano /etc/profile)
Now we can install rmr2 and rhdfs in R
Again, open R then install
> install.packages("/home/cloudera/Downloads/rmr2_3.3.1.tar.gz", type=source, repos=NULL)
> install.packages("/home/cloudera/Downloads/rhdfs_1.0.8.tar.gz", type=source, repos=NULL)If you get an error, install the package it asks for - I might have missed one or two above.
Install RStudio server.
You can follow the instructions given on the Rstudio site:
$ wget https://download2.rstudio.org/rstudio-server-rhel-0.99.467-x86_64.rpm
$ sudo yum install --nogpgcheck rstudio-server-rhel-0.99.467-x86_64.rpmRStudio server runs as a service, so it should be started immediately. You can get your ip from:
$ifconfigThen access from your host OS at http://givenip:8787
Initial Hadoop setup
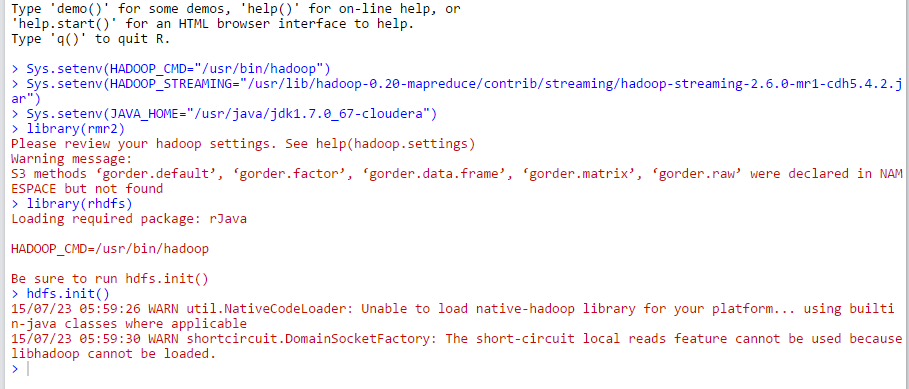
Now we can use Hadoop. In R, type the following to set where your hadoop cmd, streaming and java home are, as well as load the libraries and init the hdfs:
Sys.setenv(HADOOP_CMD="/usr/bin/hadoop")
Sys.setenv(HADOOP_STREAMING="/usr/lib/hadoop-0.20-mapreduce/contrib/streaming/hadoop-streaming-2.6.0-mr1-cdh5.4.2.jar")
Sys.setenv(JAVA_HOME="/usr/java/jdk1.7.0_67-cloudera")
library(rmr2)
library(rhdfs)
hdfs.init()There will be a couple of warnings, but hopefully no errors.
These will need to be loaded every time you want to use hadoop, it may be worth making an RProfile with them.
If you get the error which contains “Unsupported major.minor version 51.0” upon running hdfs.init() your java is set up incorrectly - make sure the java home you have defined above is version 1.7.0 or greater.
Hadoop hello world
Using the mtcars dataset, we can see what Hadoop can do:
> mtcars2 <- to.dfs(mtcars)
> count <- mapreduce(input = mtcars2,
map = function(k, v){keyval(v[2], mtcars)},
reduce = function(k, v){keyval(k,
data.frame(mpg = mean(v$mpg),
disp = mean(v$disp),
drat = mean(v$drat),
wt = mean(v$wt),
qsec = mean(v$qsec))
)})
> z <- from.dfs(count)
> as.data.frame(z)Which will give some technical info about your hadoop run:
And some data:
cyl val.mpg val.disp val.drat val.wt val.qsec
4 4 26.66364 105.1364 4.070909 2.285727 19.13727
6 6 19.74286 183.3143 3.585714 3.117143 17.97714
8 8 15.10000 353.1000 3.229286 3.999214 16.77214A mean for each variable, grouped by cylinder.
We can also do the canonical hello world for hadoop: a word count. This time on a text file of moby dick:
mobydick <- to.dfs(readLines("/home/cloudera/Downloads/mobydick.txt"))
countmapper <- function(key,line) {
word <- unlist(strsplit(line, split = " "))
keyval(word, 1)
}
z <- mapreduce(
input = mobydick,
map = countmapper,
reduce = function(k,v){keyval(k,length(v))}
)
z2<-from.dfs(z)
head(as.data.frame(z2))You should get something like this (we should strip punctuation if we want better results):
key val
1 1113
2 & 2
3 - 7
4 3 2
5 4 1
6 A 125Is this better than something you could run in dplyr in two seconds? For now, no. The beauty of rhadoop is that using mapreduce we can keep large data out of memory and run across servers on extremely large files. You should now have a system up and running where you can try out mapreduce jobs, and start thinking about analysis of big data.